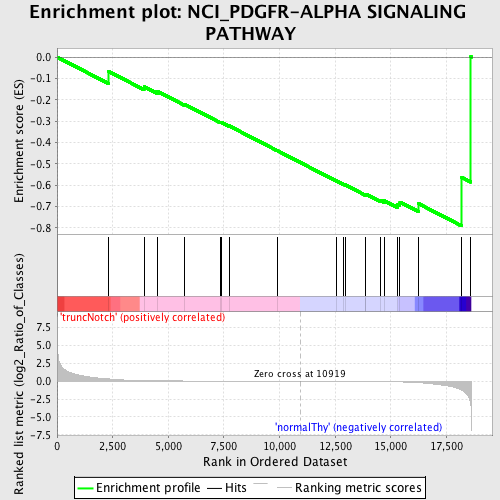
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy_repos |
| Phenotype | phenotype_truncNotch_versus_normalThy.cls#truncNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | NCI_PDGFR-ALPHA SIGNALING PATHWAY |
| Enrichment Score (ES) | -0.7905154 |
| Normalized Enrichment Score (NES) | -1.4976786 |
| Nominal p-value | 0.032407407 |
| FDR q-value | 0.38126403 |
| FWER p-Value | 0.997 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | GRB2 | 6650398 | 2319 | 0.313 | -0.0675 | No | ||
| 2 | SHB | 4920494 | 3909 | 0.080 | -0.1383 | No | ||
| 3 | CAV1 | 870025 | 4517 | 0.055 | -0.1609 | No | ||
| 4 | PDGFRA | 2940332 | 5734 | 0.028 | -0.2212 | No | ||
| 5 | PIK3R1 | 4730671 | 7321 | 0.013 | -0.3042 | No | ||
| 6 | FOS | 1850315 | 7390 | 0.012 | -0.3056 | No | ||
| 7 | SOS1 | 7050338 | 7726 | 0.010 | -0.3217 | No | ||
| 8 | PLCG1 | 6020369 | 9890 | 0.003 | -0.4375 | No | ||
| 9 | RAPGEF1 | 60040 4590670 | 12552 | -0.006 | -0.5796 | No | ||
| 10 | IFNG | 5670592 | 12891 | -0.007 | -0.5964 | No | ||
| 11 | CAV3 | 1770519 | 12943 | -0.008 | -0.5977 | No | ||
| 12 | JUN | 840170 | 13846 | -0.016 | -0.6432 | No | ||
| 13 | PIK3CA | 6220129 | 13861 | -0.016 | -0.6410 | No | ||
| 14 | ITGAV | 2340020 | 14549 | -0.033 | -0.6718 | No | ||
| 15 | CSNK2A1 | 1580577 | 14708 | -0.041 | -0.6728 | No | ||
| 16 | SHC1 | 2900731 3170504 6520537 | 15309 | -0.081 | -0.6902 | Yes | ||
| 17 | JAK1 | 5910746 | 15408 | -0.091 | -0.6788 | Yes | ||
| 18 | CRK | 1230162 4780128 | 16249 | -0.206 | -0.6863 | Yes | ||
| 19 | CRKL | 4050427 | 18189 | -1.245 | -0.5630 | Yes | ||
| 20 | SRF | 70139 2320039 | 18598 | -3.207 | 0.0010 | Yes |

